New Delhi: Scientists have developed a new technology platform to detect the Covid-19 virus by fluorescence readout. The potential of the new technology has been demonstrated for the detection of SARS-CoV-2.
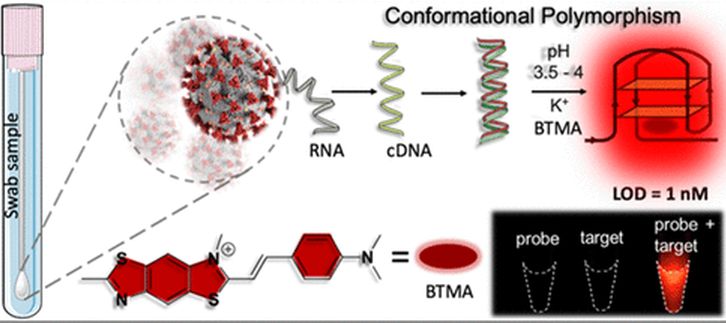
Scientists from Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR), an autonomous institute of the Department of Science & Technology, Govt. of India, along with scientists from IISc (India Institute of Science), have demonstrated a noncanonical nucleic acid-based G-quadruplex (GQ) topology targeted reliable conformational polymorphism (GQ-RCP) platform to diagnose Covid-19 clinical samples.
The present work demonstrated the first GQ-targeted diagnostic platform for SARS-CoV-2 in clinical samples, based on a novel platform GQ-RCP. This molecular detection platform can be integrated into field-deployable isothermal amplification assays with more reliability and sequence specificity.
The platform lays greater emphasis on deciphering and systematic characterization of a unique set of interactions in nucleic acids to attain stable and reliable noncanonical DNA/RNA targets. The RCP-based target validation is a general and modular approach for the development of noncanonical nucleic acid-targeted diagnostic platforms for diverse pathogens, including bacteria and DNA/RNA viruses.
RT-q-PCR has been the gold standard for accurate detection of SARS-CoV-2 (Covid-19). Among the recent innovations on a nucleic acid-targeted diagnosis of SARS-CoV-2, the techniques such as RT RPA and RT-LAMP use general-purpose DNA sensing probes. This increases the propensity of false-positive results arising out of unbiased detection of nonspecific amplification products. Recognizing unique DNA secondary conformations can be a promising solution to achieve reliable readouts. The team has identified and characterized a unique G-quadruplex-based target derived from the 30 kb (kilobytes) genomic landscape of SARS-CoV-2 for specific detection of SARS-CoV-2. Unlike the other reliable diagnostic assays where the existing fundamental concepts have been repurposed, this work presents a completely novel strategy to target a unique, unconventional structure specific to the SARS-CoV-2 sequence using small molecule fluorophores (microscopic molecules).
This technology platform can be used to detect other DNA/RNA pathogens such as HIV, influenza, HCV, Zika, Ebola, bacteria, and other mutating/evolving pathogens.